Toxic protein ‘variant’ may be the next target for ALS therapies
Study challenges conventional model of protein aggregation in neurodegenerative disease
Scientists have long known that proteins can form harmful clusters in neurodegenerative diseases like Alzheimer’s disease, Parkinson’s disease and amyotrophic lateral sclerosis (ALS). But a new study by Penn State researchers shows that ‘variant’ complexes of a protein implicated in ALS pathology form in separate pathways, a discovery which may make it easier for drug developers to design therapies to target the more harmful variant.
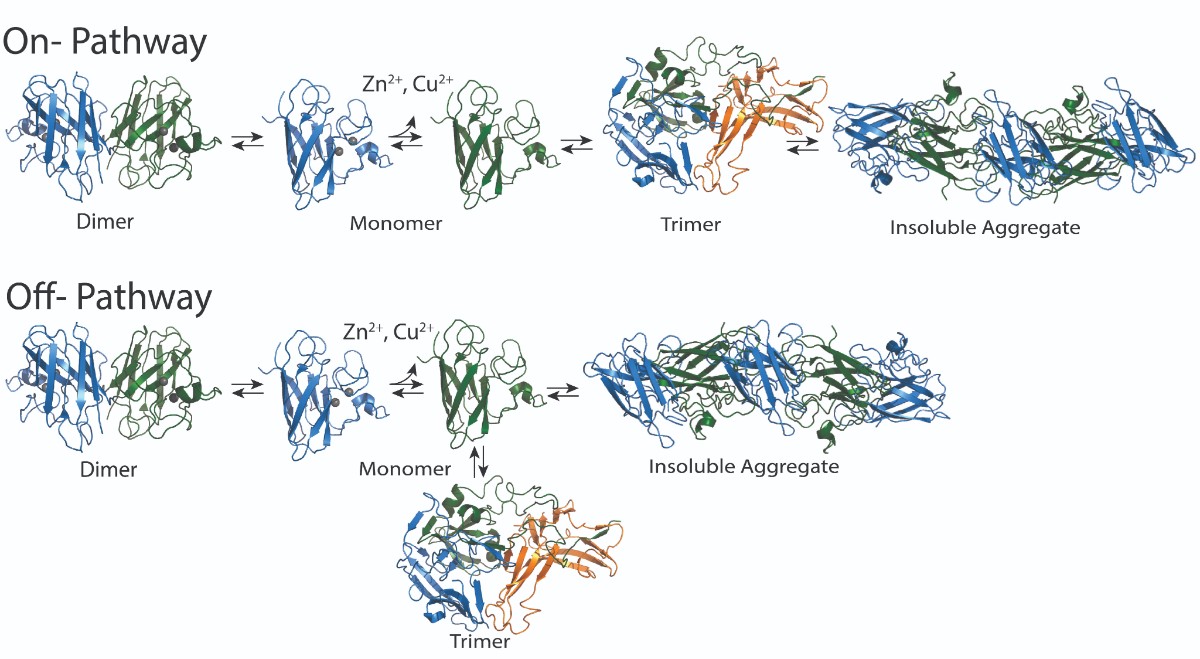
Superoxide dismutase (SOD1) is an aggregating protein that contributes to ALS development and progression, though it is unclear which biological mechanisms it uses to do this. Mutations in this protein have been implicated in 15-30% of familial ALS cases and 1-2% of spontaneous ALS cases. Normally existing in a two-part dimer, loss of SOD1 copper or zinc ions can cause it to separate into two separate monomers, or units. Monomers can form form a trimer, or three-part form, or aggregate into larger fibrils, which consist of many SOD1 monomers. Previous research showed that the trimer form is toxic to cells. Other research has suggested that the larger aggregate form may actually have a protective function.
Determining whether the trimer is needed for the formation of larger aggregates or if it is competing with the formation of aggregates could help determine a therapeutic strategy. Nikolay Dokholyan, G. Thomas Passananti Professor and vice chair for research in the Department of Pharmacology at Penn State College of Medicine , and Brianna Hnath, a graduate student in the Department of Biomedical Engineering at Penn State College of Engineering, found through their work that the trimer develops in a separate, competitive path from the larger aggregates.
“Determining whether trimer formation is separate from, or part of, large aggregate development could affect the way we pharmaceutically treat diseases like ALS,” Hnath said. “Current therapeutics focus on breaking down the large aggregates, but in situations where aggregates turn out to be protective, it may be better to instead focus on targeting the trimer formation.”
Hnath and Dokholyan designed stable SOD1 trimer mutants and using neuron-like cell models, found that stabilized SOD1 trimers increased cell death. They also determined that stabilizing the trimers prevented fibril formation in the cell models. This observation led them to perform chromatography experiments where they found that SOD1 trimers directly compete with fibrils – large complexes of SOD1 monomers.
Most importantly, the researchers determined that the trimer structure forms independently of the larger aggregates, which confirmed that trimer formation is not part of the pathway to building of SOD1 fibril aggregates. The results were published in the Biophysical Journal.
“This finding is significant because it shows us that therapeutic strategies for ALS can focus on inhibiting the formation of toxic trimers, as opposed to targeting the fibrillar aggregates, like some medications for Alzheimer’s do,” said Dokholyan, who is also a professor of biochemistry and molecular biology, biomedical engineering, chemistry and a researcher with Penn State Cancer Institute and Penn State Neuroscience Institute. “The fact that smaller oligomers are toxic in ALS is a relatively newer theory that is still needs further exploration and validation.”
Dokholyan and Hnath plan to conduct studies to better understand the structure of the SOD1 trimer and how protein-protein interactions related to the trimer contribute to ALS development and progression.
This research was supported by the National Institutes of Health and the Passan Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funders. The researchers declare no conflicts of interest.
If you're having trouble accessing this content, or would like it in another format, please email Penn State Health Marketing & Communications.
