Researchers secure $3.3M NIH grant to create 3D zebrafish atlas
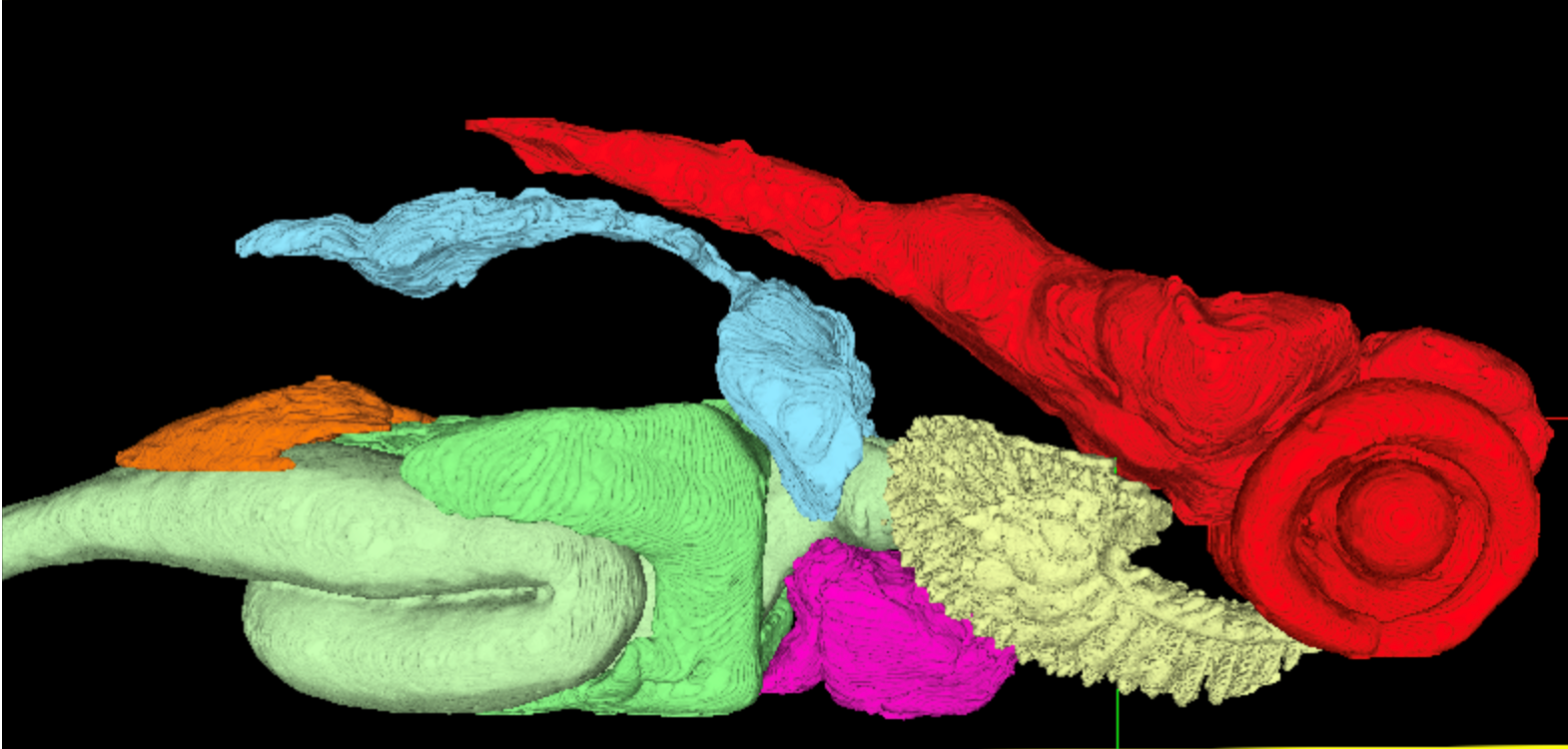
Researchers at Penn State College of Medicine have been awarded a $3.3 million grant from the National Institutes of Health (NIH) Office of Research Infrastructure Programs (ORIP) to develop an integrated 3D digital microanatomical atlas of the zebrafish. This project will provide an open-access resource that integrates high-resolution 3D anatomical data with genomic information to facilitate research for understanding human diseases and drug discovery.
Dr. Khai C. Ang, assistant professor of pathology and laboratory medicine and the scientific director of Penn State Zebrafish Functional Genomics Facility, will lead the four-year, multi-institutional project titled “3D Zebrafish Microanatomical and Gene Expression Atlas for Disease Modeling.” His collaborators include Dr. John Postlethwait, professor of biology at the University of Oregon, and Dr. Keith Cheng, distinguished professor of pathology at the College of Medicine.
The research team will create a comprehensive, web-based atlas that maps the intricate organization of cells and tissues in zebrafish and their respective gene expression profiles at various life stages. By combining cutting-edge imaging techniques like X-ray 3D histotomography with detailed gene expression data, the atlas will offer a unique, multidimensional view of a whole vertebrate organism.
“A significant challenge in biology is understanding how the vast amount of genomic data we have relates to the phenotypes of an organism,” said Dr. Ang. “This atlas will bridge that gap, allowing researchers to see not only where specific genes are active, but also to understand their function within the 3D context of the entire animal. This will be a transformative tool for the research community.”
Zebrafish are a powerful model organism for studying human biology and disease. They share 70 percent of their genes with humans and 84 percent of genes known to be associated with human disease have a zebrafish counterpart. Their rapid development and transparent embryos make them ideal for observing biological processes in real-time. This project will leverage these advantages to create a resource with broad applications across many fields of study, including cancer, neurodegenerative diseases and developmental disorders.
The project centers on two innovations:
- Next-Generation 3D Imaging: Using a high-resolution, wide-field 3D x-ray imaging technique called histotomography, the team will generate detailed, “sliceable” digital models of zebrafish. This provides a complete, undistorted view of its internal structures, a significant leap forward from traditional 2D histology, which involves physical slicing that damages the tissue. Dr. Cheng, through a separate NIH grant, and his team are building infrastructure to enable 3D X-ray histotomography access at synchrotron beamlines for biology.
- “Omics” Integration: The atlas will integrate transcriptomic data, which provides a snapshot of all the gene activity within the fish. This will allow scientists to connect molecular-level changes to anatomical features, providing a more holistic understanding of health and disease. Dr. Postlethwait and his team will be generating spatial-transcriptomic data for the project.
The resulting open-access, web-based atlas will be created with input from the broader zebrafish research community to ensure it remains a dynamic and interactive resource for scientists worldwide. Researchers will be able to compare their own experimental data with the reference atlas, accelerating their research and fostering collaboration.
“This project is not just about creating a static map; it’s about building a dynamic and collaborative platform,” added Dr. Ang. “By providing this resource to the scientific community, we hope to empower new discoveries that will ultimately improve human health.”
If you're having trouble accessing this content, or would like it in another format, please email Penn State Health Marketing & Communications.
