College of Medicine shares core facility publication highlights
Penn State College of Medicine’s core facilities and research resources have provided support to enable researchers to produce more than 100 publications from 2019 through mid-2020, including the ones featured here.
See a larger list of publications citing the cores here
Cores used: Advanced Light Microscopy, Flow Cytometry, Genome Sciences
Dhoonmoon A, Schleicher EM, Clements KE, Nicolae CM, Moldovan G-L. Genome-wide CRISPR synthetic lethality screen identifies a role for the ADP-ribosyltransferase PARP14 in DNA replication dynamics controlled by ATR. Nucleic Acids Res. 2020 July 27;48(13):7252-7264.
This work used multiple cores to identify an unexpected role for the ATR-CHK1 pathway in promoting cellular viability in the context of PARP14 deficiency. This indicates that the status of the PARP14 gene in tumors is an important determinant of the tumor response to DNA damage repair (DDR) inhibitors, which are emerging as a powerful class of cancer drugs.
Cores used: Flow Cytometry, Genome Sciences
Chodisetti SB, Fike AJ, Domeier PP, Singh H, Choi NM, Corradetti C, Kawasawa YI, Cooper TK, Caricchio R, Rahman ZSM. Type II but not Type I IFN signaling is indispensable for TLR7-promoted development of autoreactive B cells and systemic autoimmunity. J Immunol. 2020; 204:796-809; doi: 10.4049/jimmunol.1901175.
This work used flow cytometric analysis and RNASeq transcriptomic analysis to define a previously unrecognized and indispensable role for IFN-γ signaling, as well as its downstream transcription factors STAT1 and T-bet, in B cells in promoting TLR7-accelerated antibody production in both extrafollicular Ab-forming cell and follicular germinal center responses in the development of autoimmunity in systemic lupus erythematosus.
Cores Used: Bioluminescence Imaging, shRNA Core, Advanced Light Microscopy Imaging
Kim YS, Gupta-Vallur P, Jones VM, Worley BL, Shimko S, Shin DH, Crawford LC, Chen CW, Aird KM, Abraham T, Shepherd TG, Warrick JI, Nam NY, Phaeton R, Mythreye K, Hempel N. Context-dependent activation of SIRT3 is necessary for anchorage-independent survival and metastasis of ovarian cancer cells. Oncogene. 2019 Nov 13. doi:10.1038/s41388-019-1097-7.
This research demonstrated the importance for peritoneal tumor formation (from injected ovarian cancer cells cultured from human patient ascites fluid) of both the deacetylase/sirtuin SIRT3, as well as superoxide dismutase SOD2. Inhibition of those enzyme activities, with either shRNA or siRNA, significantly decreased tumor establishment, as followed serially in the same animals for 28 days using bioluminescence imaging and as imaged by 3D multiphoton imaging of tumor cell spheroids.
Cores used: Advanced Light Microscopy Imaging, Electron Microscopy, Flow Cytometry
Tang Z, Takahashi Y, He H, Hattori T, Chen C, Liang X, Chen H, Young MM, Wang HG. TOM40 targets Atg2 to mitochondria-associated ER membranes for phagophore expansion. Cell Rep. 2019 Aug 13;28(7):1744-1757.e5. doi:10.1016/j.celrep.2019.07.036.
This work used correlative light and electron microscopy, electron microscopy, confocal microscopy and flow cytometry to help show that TOM40 recruits mammalian Atg2A as a critical regulator of Atg9A vesicle delivery and phagophore expansion during autophagosome biogenesis at the mitochondrial associated endoplasmic reticulum membrane. This process is critical for macroautophagy, an intracellular degradation process that plays a vital role in the maintenance of cellular and tissue homeostasis, as well as in the pathogenesis of various human diseases, including cancer, vascular and neurodegenerative disease.
Cores used: Genome Sciences
Dierschke SK, Miller WP, Favate JS, Shah P, Imamura Kawasawa Y, Salzberg AC, Kimball SR, Jefferson LS, Dennis MD. O-GlcNAcylation alters the selection of mRNAs for translation and promotes 4E-BP1-dependent mitochondrial dysfunction in the retina. J Biol Chem. 2019 Apr 5;294(14):5508-5520.
This research used RNA transcriptomics and polyribosome-mRNA profiling to show that diabetes-induced O-linked glycosylation (O-GlcNAcylation) alters mRNA translation in a manner that causes mitochondrial oxidative stress in the retina. The research also showed that while enhanced O-GlcNAcylation only changed mRNA transcript abundance of ~1% of all mRNAs, it altered the polyribosome-associated mRNAs (those that are actually being translated) of approximately 19% of the whole transcriptome. This is consistent with a much larger role of translational control than of transcriptional control (mRNA abundance) changes on gene protein expression levels.
Cores used: Mass Spectrometry and Proteomics
Huang X, Sterling NW, Du G, Sun D, Stetter C, Kong L, Zhu Y, Neighbors J, Lewis MM, Chen H, Hohl RJ, Mailman RB. Brain cholesterol metabolism and Parkinson’s disease. Mov Disord. 2019;34(3):386-395.
This research used mass spectrometry to measure the fasting plasma levels of two low-abundance cholesterol metabolites, (S)24‐OH‐cholesterol (brain‐derived cholesterol metabolite) and 27‐OH‐cholesterol (peripheral cholesterol metabolite), from 60 Parkinson’s disease patients and 64 control patients, with follow-up 36 months later of 35 Parkinson’s disease patients and 30 control patients. These results indicate that plasma (S)24‐OH‐cholesterol (possibly reflecting brain cholesterol metabolism) is inversely linked to Parkinson’s disease, is relatively stable over time and may serve as a new biomarker for Parkinson’s disease.
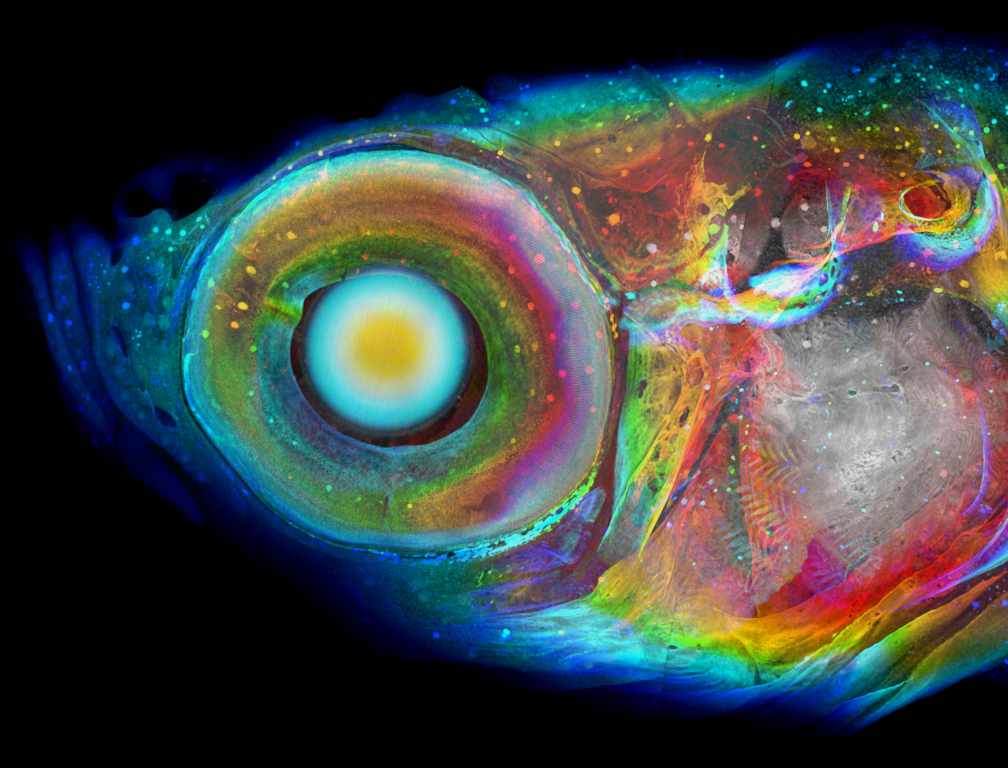
Cores used: Zebrafish Functional Genomics Core
Ding Y, Vanselow DJ, Yakovlev MA, Katz SR, Lin AY, Clark DP, Vargas P, Xin X, Copper JE, Canfield VA, Ang KC, Wang Y, Xiao X, De Carlo F, van Rossum DB, La Riviere P, Cheng KC. Computational 3D histological phenotyping of whole zebrafish by X-ray histotomography. eLife. 2019;8:e44898.
This new method of 3D whole-zebrafish histotomography with X-ray synchrotron microCT demonstrated that one could compare the whole-body phenotypic effects of different individual gene mutations at the level of individual cells throughout the entire body of juvenile zebrafish, including soft tissues not amenable to histological examination. These accurate measurements of microscopic features should allow similar examination of phenotypic effects of other gene mutations, as well as drug and environmental toxin exposures.
Read more
- Explore research at Penn State College of Medicine
- Search the Pure researcher directory
- See a list of NIH and other grants received by the college
- Learn about the college’s core facilities
- Read more College of Medicine research news
If you're having trouble accessing this content, or would like it in another format, please email the Penn State College of Medicine web department.
