New computational tool allows researchers to predict key functional sites in proteins
A new technology that uses a protein’s structure to predict the inner wiring that controls the protein’s function and dynamics is now available for scientists to utilize. The tool, developed by researchers at Penn State, may be useful for protein engineering and drug design.
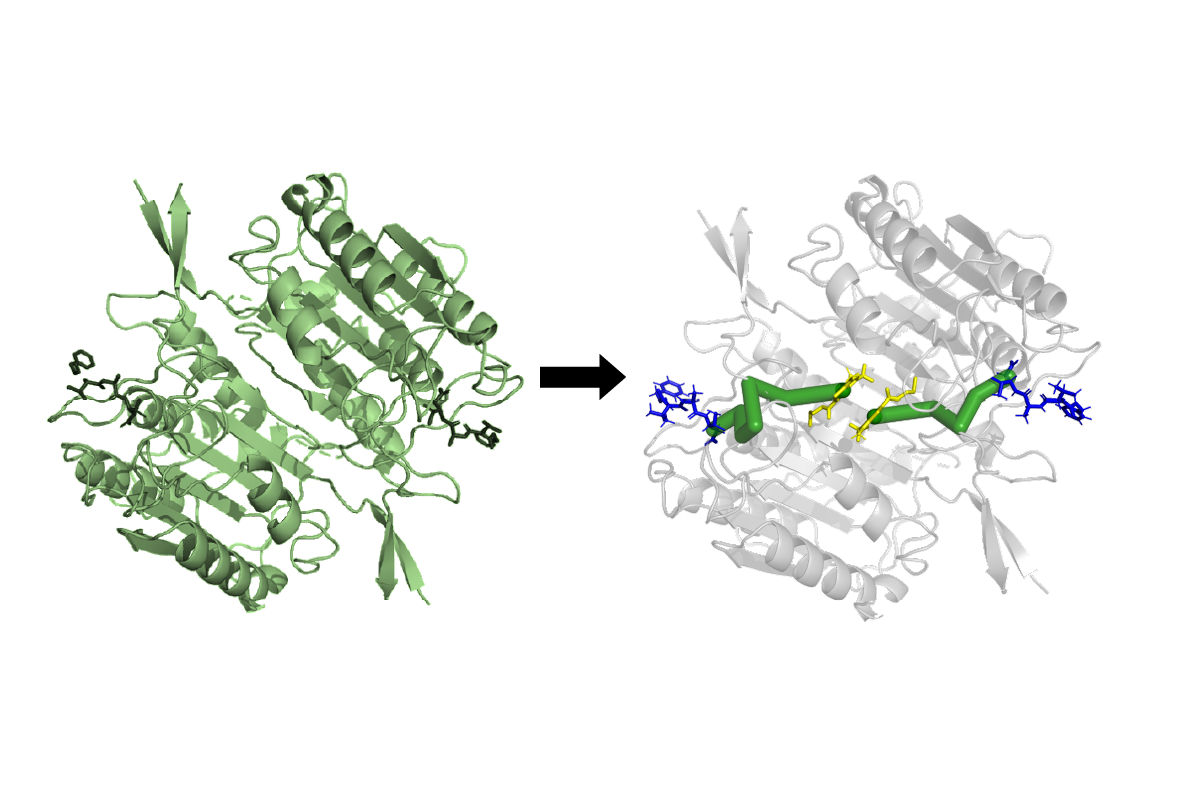
Nikolay Dokholyan, professor of pharmacology at Penn State College of Medicine, and postdoctoral scholar Jian Wang created an algorithm called Ohm that can predict allosteric sites in a protein. These are locations where proteins are particularly sensitive to relay certain changes in their structure and function as a result of external stimuli including other proteins, small molecules, water or ions. Signaling at and between allosteric sites in proteins regulate many biological processes.
According to Dokholyan, Ohm’s ability to predict allosteric sites in proteins may be useful for developing targeted therapeutics for certain disease states. He said that many drugs on the market, such as G Protein-Coupled Receptor (GPCR) drugs, may cause unintended side effects because they target proteins that are similar in structure to their intended target.
“Drugs designed to target specific allosteric sites on a protein of interest can hopefully avoid side effects caused by drugs that target similar proteins,” Dokholyan said. “Ohm may be useful for biomedical researchers seeking to identify allosteric sites in proteins that play key roles in biological processes of certain diseases.”
Proteins carry out essential functions in the body and are built using genetic code inscribed in a person’s DNA. Each protein is built using sequences of 20 different amino acids. Wang and Dokholyan hypothesized that the physical forces from interactions between the atoms that make up the amino acids would allow them to predict allosteric pathways and sites in proteins. Ohm was designed to account for the interactions between atoms and identifies areas of density in proteins to predict allosteric pathways and sites in proteins.
“In a crystalline structure, atoms are spaced evenly apart and energy flows through it in an even fashion,” Dokholyan said. “Proteins’ structures are heterogeneous, so energy will flow through them in regions where the atoms are more densely packed together. Ohm identifies regions and pathways of atomic density that allow it to predict allosteric sites in proteins.”
They tested the functionality of the program by inputting the genetic data from 20 proteins with known allosteric sites to see if the program would accurately predict the same spots. Results from the analysis, published in Nature Communications, showed that Ohm identified many of the same allosteric sites as those predicted from previous methods and experiments.
Dokholyan, a member of Penn State Cancer Institute, said that Ohm can analyze allosteric paths in any protein and that researchers can access the tool through dokhlab.med.psu.edu/ohm.
“Researchers around the world can use Ohm to predict allosteric sites and pathways in their protein of interest,” Wang said. “This tool will be essential for the future of allosteric drug development that seeks to reduce unwanted side effects through specific targeting.”
Abha Jain, Leanna McDonald, Craig Gambogi and Andrew Lee of the University of North Carolina at Chapel Hill also contributed to this research.
The authors declare no conflict of interest.
The project described was supported by the Passan Foundation, the National Institutes of Health and Penn State Clinical and Translational Science Institute via the National Center for Advancing Translational Sciences, through Grant UL1 TR002014. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
If you're having trouble accessing this content, or would like it in another format, please email Penn State Health Marketing & Communications.
