New way to image whole organisms in 3D brings key skin color pigment into focus
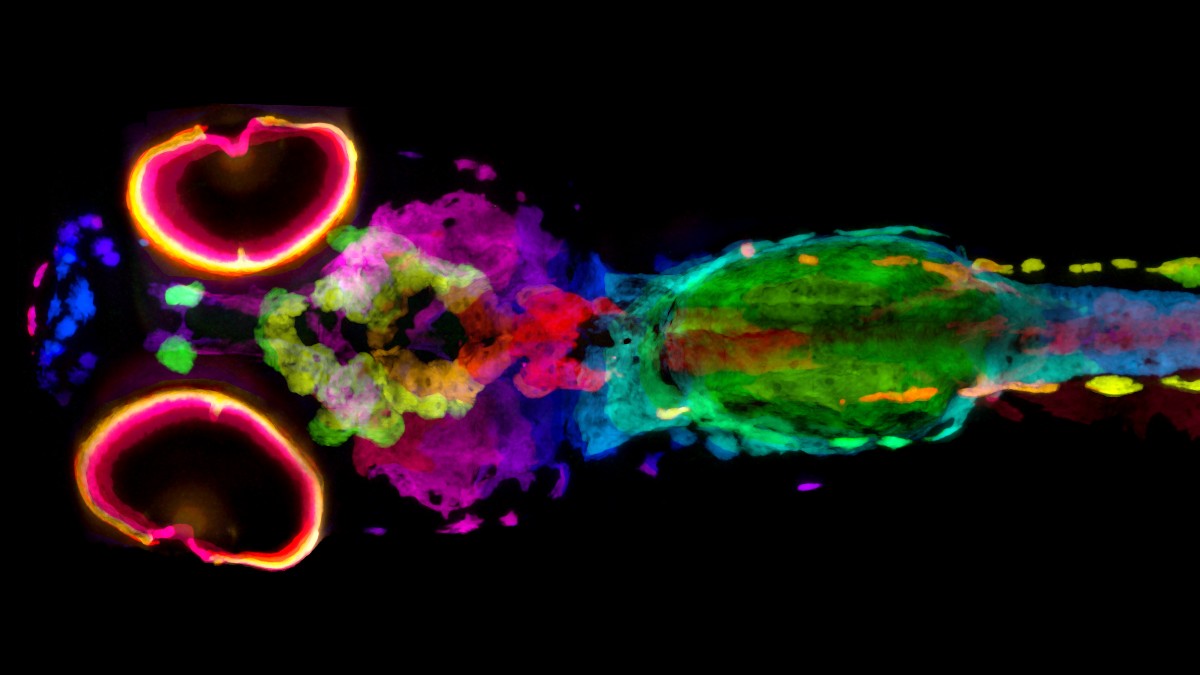
To understand the biological underpinnings of skin and hair pigmentation and related diseases such as albinism or melanoma, scientists and doctors need quantitative, three-dimensional information about the architecture, content and location of pigment cells. Penn State College of Medicine researchers have developed a new technique that allows scientists to visualize every cell containing melanin pigment in 3D, in whole zebrafish.
Studying melanin is challenging because it blocks the light used in traditional microscopy. So the researchers turned to X-ray imaging, which can pass through optically opaque matter like melanin.
In 2019, a team led by Dr. Keith Cheng, distinguished professor of pathology, pharmacology and biochemistry and molecular biology, developed “X-ray histotomography” — a cellular form of CT imaging, as a method for investigating the 3D architecture of cells and tissues in biological samples at unprecedented resolution and clarity. Spencer Katz, an MD/PhD medical scientist training program student on Cheng’s team, has modified this micro-CT technique to specifically investigate melanin, a pigment scientists are studying in human skin color and melanoma research, in whole zebrafish.
Melanin is a brown to black pigment that gives the zebrafish its characteristic stripes and humans dark skin, hair and eyes. More than 15 years ago, Cheng and his lab discovered a key gene in the evolution of light skin color in humans by studying a particular mutant zebrafish line, golden, that has lighter stripes. This discovery demonstrated the relevance of zebrafish models for studying critical questions about human biology and disease, including albinism and melanoma.
Micro-CT, like human CT, uses a series of X-rays taken at slightly different angles to compute, or reconstruct, 3D representations of the original object. For micro-CT, the samples are smaller and the resolution developed by Cheng’s team is 2000-fold higher. Katz used silver to stain the melanin, which allowed the researchers to determine the 3D location and density of melanin from scans of whole zebrafish.
To perform the imaging, the Cheng Lab partnered with Dilworth Parkinson at the Advanced Light Source at the Lawrence Berkeley National Labs in Berkeley, California — home of one of America’s most powerful synchrotron X-ray sources, where he directs a micro-CT resource suitable for Cheng’s X-ray histotomography. The lab’s new X-ray detector system was designed to achieve resolutions unprecedented for samples the size of whole zebrafish or human biopsies. The team scanned zebrafish with both normal and altered pigmentation, including golden.
The researchers were able to visualize every cell containing melanin, called melanocytes, in the fish, and map each of their positions in 3D. Moreover, they could get quantitative measurements of their melanin content, allowing direct comparison of melanin content across normal and mutant fish for the first time. They published the results of their study in eLife.
This work laid a foundation for further research on melanin-containing cancers, or melanomas, which are typically graded by the depth of tumor cell invasion. According to Cheng, a researcher at Penn State Cancer Institute, a number of zebrafish models of melanoma exist and can be studied using the new technique. Katz and Cheng said that human melanomas can be stained with silver and imaged in the same way to more completely characterize the tumor cells and their arrangements in tumors. They predict that scientists will be able to, for example, count the number of tumor cells of different characteristics and more definitively study invasion, a central characteristic of cancer, helping doctors with prognostic and treatment decisions.
In the future, the Cheng Lab will continue to develop new staining and optical methods for expanding the applications of histotomography. This study represents a proof-of-principle for how whole-body, 3D computational analysis of organisms and tissues can be achieved using micro-CT that may allow a far more complete understanding of gene function.
Maksim Yakovlev, Daniel Vanselow, Yifu Ding, Alex Lin, Victor Canfield and Khai Chung Ang of Penn State College of Medicine also contributed to this research. The new lens system used in the study was designed and built by Yuxin Wang of Mobile Imaging Innovations, Inc. The authors declared no competing interests.
This research was supported by the National Institutes of Health, the Pennsylvania Department of Health Tobacco CURE Funds, the Jake Gittlen Laboratories for Cancer Research at Penn State College of Medicine, Penn State Huck Institutes of the Life Sciences and Penn State Institute for Computational and Data Sciences.
This study utilized the College of Medicine’s zebrafish functional genomics core and represents activities of the Penn State Center for Artificial Intelligence Foundations and Scientific Applications (CENSAI) and computational phenomics initiatives.
If you're having trouble accessing this content, or would like it in another format, please email Penn State Health Marketing & Communications.
